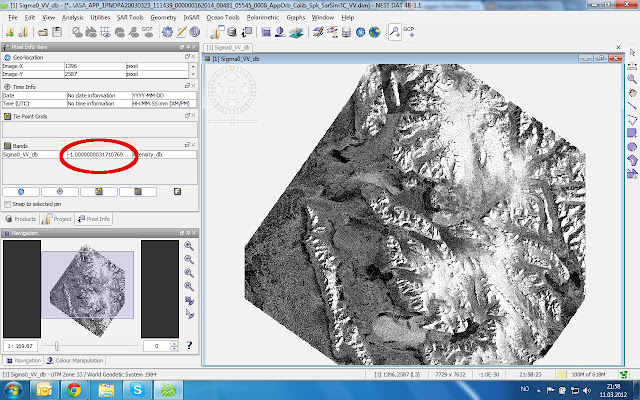
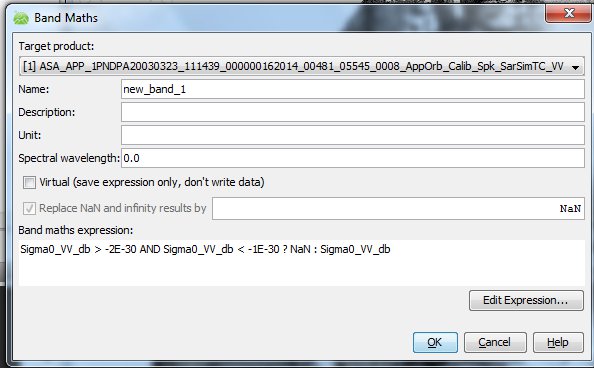
The Band Math uses a C-code If-expression which in this case says: If the image value is between -1E-30 and -2E-30, then replace it by NaN, otherwise keep the original value.
The following is not very clear to me -- You have to deselect "virtual" and the result is written into a new channel (save your file afterwards).
It would seem that you then simply could delete the original channel and retain the new band, but that does not seem to work, somehow the new band is still referenced to the original one. You have to choose Utilities>Spatial Subset from View to do the job. Here I go straight to the "Band Subset" tab and deselect the original band:
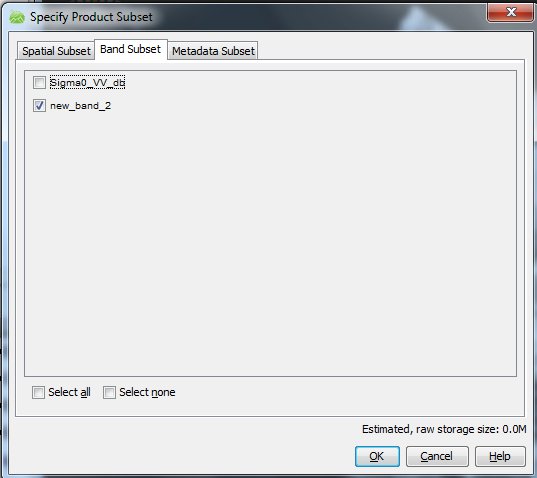
If I know choose "ok", you get a new product with the one corrected channel, which you then can save to disc.
This is of course too tedious if you have dozens of images, so to do this in batch mode, I create the processing line in "Graph Builder"
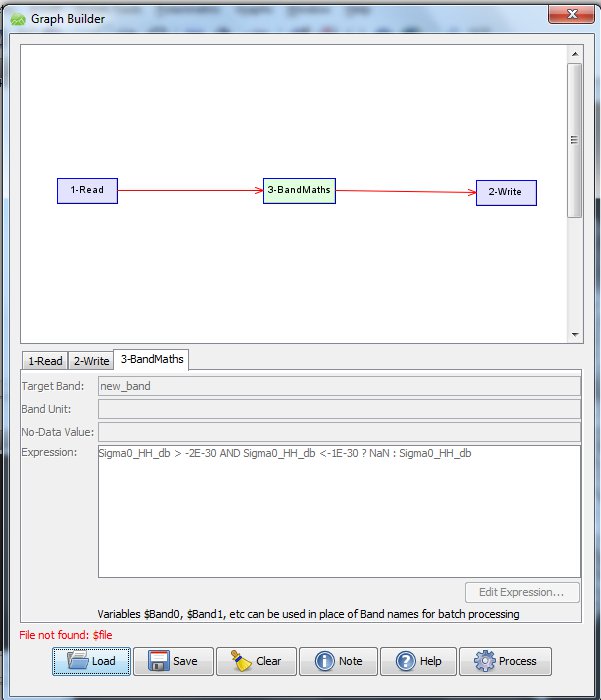
Then I save this graph as an XML file. You edit this XML-file so that the input and output file names have a place holder "$file" and "$target". Look in 1-Read that it reads "<file>$file</file>" and in 2-Write that it reads "<file>$target</file>".
Now you create a RemoveOutliers.bat file containing
for /r C:Usersmaxlocation_of_files %%X in (*.dim) do (gpt C:Usersmaxlocation_of_XMLfile RemoveOutliers.xml -Pfile="%%X" -Tfile=" C:Usersmaxlocation_of_new_files\%%~nX.dim")
What happens here?
- The for-command goes through the directory containing your files to find files named "*.dim" and passes the file name to "%%X".
- For each of these input files "-Pfile="%%X", the NEST command "gpt" applies the Graph Builder production chain saved in "RemoveOutliers.xml"
- The output is saved in the parameter -Tfile, which here is written "%%~nX.dim", the same filename but in a new directory (but you also can give it a new name like "%%~nX_NaN.dim" if you wish)
You navigate the DOS window (type "cmd" at Windows Start> "search programs and files" to open it) to the directory containing RemoveOutliers.bat, then type "RemoveOutliers.bat" and all scenes in the specified folder will be processed.
Curiously, and conveniently, if you run Band Math using "gpt" from commandline, the new file only contains the new band, so no reason for manually deleting the original band as in the NEST GUI version above is needed.
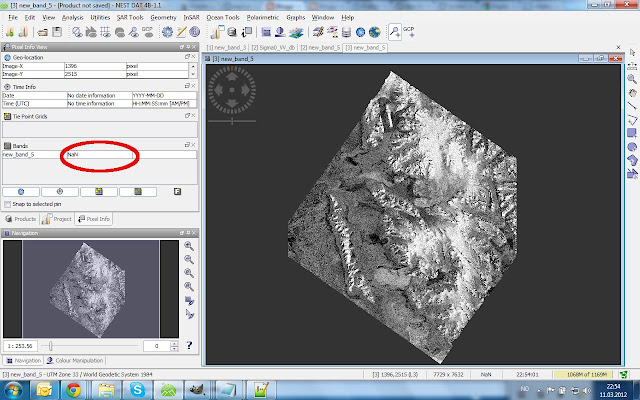
The Result (what is dark outside the scene is now NaN)
Curiously, and conveniently, if you run Band Math using "gpt" from commandline, the new file only contains the new band, so no reason for manually deleting the original band as in the NEST GUI version above is needed.
The Result (what is dark outside the scene is now NaN)
